| Home | |||
| News | |||
| Introduction | |||
| Methods | |||
| Flow Chart | |||
| References | |||
| Download | |||
| About | |||
| FAQ | |||
| Search | |||
|
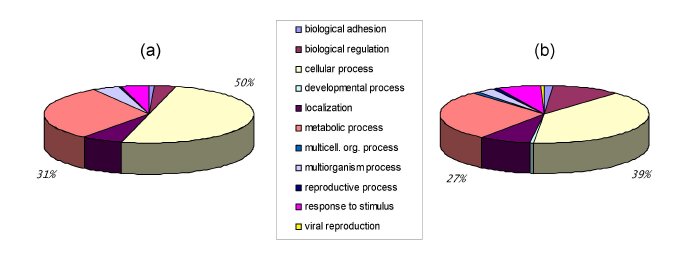
Description of the LigASite gold-standard datasetLigASite is a gold-standard dataset of biologically relevant binding sites in protein structures. It consists of proteins with one unbound structure and at least one structure of the protein-ligand complex. Both a redundant and a non-redundant (sequence identity lower than 25%) version is available. Quaternary structures proposed by PISA (3) are used for all structures in the dataset. A common problem when automatically deriving datasets of binding sites in protein structures is that ligands which appear in PDB files are often contaminants that do not bind specifically to the protein. In order to automatically filter out these biologically irrelevant ligands from our dataset, we use a procedure that considers the number of heavy atoms (i.e. non-hydrogen atoms) in the ligand and the number of inter-atomic contacts between the protein and the ligand. The underlying assumptions used are that (i) the proportion of biologically irrelevant ligands is larger for smaller molecules, and (ii) ligands that bind in biologically important sites (whether they be the ligand that occurs in vivo or an analogue of it) should generally interact specifically with the proteins and should therefore make more inter-atomic contacts with these. Inter-atomic contacts are identified from the three-dimensional structures with the program LPC (5). The availability of both unbound and bound structures for each protein guarantees that our dataset can be used to benchmark binding site prediction methods, in conditions that mimic cases where the binding site is truly unknown. In cases where several different bound structures are available for a given protein, all are used to define the binding sites. As it relies on simple numeric cutoffs, the LigASite dataset can be automatically updated as new data become available in the PDB (2). The fact that the construction of the dataset does not depend on external annotation systems and databases also ensures optimal representativity of the data contained in the PDB. The pie graph below shows that the distribution of functions present in LigASite do not differ substantially from the distribution of functions in a non-redundant version of the PDB.
The non-redundant version of the dataset is available here. The redundant version of the dataset is available here. A specific page is available for each protein in the dataset and can be accessed from the dataset front page. A precise description of the steps to construct the dataset is available here. |
April 2012
University College London, Biomolecular Structure and Modelling Unit, London, UK
Hospital for Sick Children and University of Toronto, Structural Biology and Biochemistry Program, Toronto, Canada