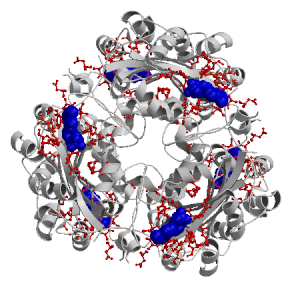
| ANP F 502 |
|---|
| 15 | D |
LYS |
| 55 | D |
TYR |
| 57 | D |
GLU |
| 58 | D |
HIS |
| 61 | D |
LYS |
| 63 | D |
PHE |
| 67 | D |
LEU |
| 70 | D |
TYR |
| 91 | D |
ARG |
| 97 | D |
THR |
| 108 | D |
ARG |
| 117 | D |
ASP |
| 118 | D |
ALA |
| 119 | D |
ILE |
| | 121 | D |
ASN |
| 124 | D |
HIS |
| 127 | D |
ASP |
| ANP X 502 |
|---|
| 15 | V |
LYS |
| 55 | V |
TYR |
| 57 | V |
GLU |
| 58 | V |
HIS |
| 61 | V |
LYS |
| 63 | V |
PHE |
| 67 | V |
LEU |
| 70 | V |
TYR |
| 91 | V |
ARG |
| 97 | V |
THR |
| 108 | V |
ARG |
| | 117 | V |
ASP |
| 118 | V |
ALA |
| 119 | V |
ILE |
| 121 | V |
ASN |
| 124 | V |
HIS |
| 127 | V |
ASP |
| ANP R 502 |
|---|
| 15 | P |
LYS |
| 55 | P |
TYR |
| 57 | P |
GLU |
| 58 | P |
HIS |
| 61 | P |
LYS |
| 63 | P |
PHE |
| 67 | P |
LEU |
| 70 | P |
TYR |
| | 91 | P |
ARG |
| 97 | P |
THR |
| 108 | P |
ARG |
| 117 | P |
ASP |
| 118 | P |
ALA |
| 119 | P |
ILE |
| 121 | P |
ASN |
| 124 | P |
HIS |
| 127 | P |
ASP |
| ANP L 502 |
|---|
| 15 | J |
LYS |
| 55 | J |
TYR |
| 57 | J |
GLU |
| 58 | J |
HIS |
| 61 | J |
LYS |
| | 63 | J |
PHE |
| 67 | J |
LEU |
| 70 | J |
TYR |
| 91 | J |
ARG |
| 97 | J |
THR |
| 108 | J |
ARG |
| 117 | J |
ASP |
| 118 | J |
ALA |
| 119 | J |
ILE |
| 121 | J |
ASN |
| 124 | J |
HIS |
| 127 | J |
ASP |
| ANP G 501 |
|---|
| 15 | E |
LYS |
| 55 | E |
TYR |
| | 57 | E |
GLU |
| 58 | E |
HIS |
| 61 | E |
LYS |
| 62 | G |
PRO |
| 63 | E |
PHE |
| 67 | E |
LEU |
| 70 | E |
TYR |
| 85 | M |
TYR |
| 91 | E |
ARG |
| 97 | E |
THR |
| 100 | G |
LYS |
| 108 | E |
ARG |
| 118 | E |
ALA |
| 119 | E |
ILE |
| 121 | E |
ASN |
| | 124 | E |
HIS |
| 127 | E |
ASP |
| ANP I 501 |
|---|
| 15 | G |
LYS |
| 55 | G |
TYR |
| 57 | G |
GLU |
| 58 | G |
HIS |
| 61 | G |
LYS |
| 62 | E |
PRO |
| 63 | G |
PHE |
| 67 | G |
LEU |
| 70 | G |
TYR |
| 85 | Y |
TYR |
| 91 | G |
ARG |
| 97 | G |
THR |
| | 100 | E |
LYS |
| 108 | G |
ARG |
| 118 | G |
ALA |
| 119 | G |
ILE |
| 121 | G |
ASN |
| 124 | G |
HIS |
| 127 | G |
ASP |
| ANP A 501 |
|---|
| 15 | Y |
LYS |
| 55 | Y |
TYR |
| 57 | Y |
GLU |
| 58 | Y |
HIS |
| 61 | Y |
LYS |
| 62 | A |
PRO |
| 63 | Y |
PHE |
| | 67 | Y |
LEU |
| 70 | Y |
TYR |
| 85 | G |
TYR |
| 91 | Y |
ARG |
| 97 | Y |
THR |
| 100 | A |
LYS |
| 108 | Y |
ARG |
| 118 | Y |
ALA |
| 119 | Y |
ILE |
| 121 | Y |
ASN |
| 124 | Y |
HIS |
| 127 | Y |
ASP |
| ANP C 501 |
|---|
| 15 | A |
LYS |
| 55 | A |
TYR |
| | 57 | A |
GLU |
| 58 | A |
HIS |
| 61 | A |
LYS |
| 62 | Y |
PRO |
| 63 | A |
PHE |
| 67 | A |
LEU |
| 70 | A |
TYR |
| 85 | S |
TYR |
| 91 | A |
ARG |
| 97 | A |
THR |
| 100 | Y |
LYS |
| 108 | A |
ARG |
| 118 | A |
ALA |
| 119 | A |
ILE |
| 121 | A |
ASN |
| | 124 | A |
HIS |
| 127 | A |
ASP |
| ANP J 502 |
|---|
| 15 | H |
LYS |
| 55 | H |
TYR |
| 57 | H |
GLU |
| 58 | H |
HIS |
| 61 | H |
LYS |
| 63 | H |
PHE |
| 67 | H |
LEU |
| 70 | H |
TYR |
| 91 | H |
ARG |
| 97 | H |
THR |
| 108 | H |
ARG |
| 117 | H |
ASP |
| | 118 | H |
ALA |
| 119 | H |
ILE |
| 121 | H |
ASN |
| 124 | H |
HIS |
| 127 | H |
ASP |
| ANP O 501 |
|---|
| 15 | M |
LYS |
| 55 | M |
TYR |
| 57 | M |
GLU |
| 58 | M |
HIS |
| 61 | M |
LYS |
| 62 | S |
PRO |
| 63 | M |
PHE |
| 67 | M |
LEU |
| 70 | M |
TYR |
| | 85 | E |
TYR |
| 91 | M |
ARG |
| 97 | M |
THR |
| 100 | S |
LYS |
| 108 | M |
ARG |
| 118 | M |
ALA |
| 119 | M |
ILE |
| 121 | M |
ASN |
| 124 | M |
HIS |
| 127 | M |
ASP |
| ANP U 501 |
|---|
| 15 | S |
LYS |
| 55 | S |
TYR |
| 57 | S |
GLU |
| 58 | S |
HIS |
| | 61 | S |
LYS |
| 62 | M |
PRO |
| 63 | S |
PHE |
| 67 | S |
LEU |
| 70 | S |
TYR |
| 85 | A |
TYR |
| 91 | S |
ARG |
| 97 | S |
THR |
| 100 | M |
LYS |
| 108 | S |
ARG |
| 118 | S |
ALA |
| 119 | S |
ILE |
| 121 | S |
ASN |
| 124 | S |
HIS |
| 127 | S |
ASP |
| | ANP D 502 |
|---|
| 15 | B |
LYS |
| 55 | B |
TYR |
| 57 | B |
GLU |
| 58 | B |
HIS |
| 61 | B |
LYS |
| 63 | B |
PHE |
| 67 | B |
LEU |
| 70 | B |
TYR |
| 91 | B |
ARG |
| 97 | B |
THR |
| 108 | B |
ARG |
| 117 | B |
ASP |
| 118 | B |
ALA |
| 119 | B |
ILE |
| | 121 | B |
ASN |
| 124 | B |
HIS |
| 127 | B |
ASP |
|