 |
LigASite database of binding sites |
|
PDB ID and HEADER, TITLE and
COMPND records of the PDB file. | | (click anywhere in this window to remove it) |
|
| 1rxs |
|
|
| E. COLI URIDINE PHOSPHORYLASE: 2'-DEOXYURIDINE PHOSPHATE COMPLEX |
TITLE |
|
|
| URIDINE PHOSPHORYLASE |
COMPND |
|
|
|
|
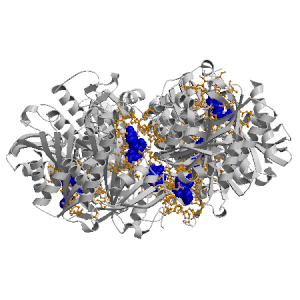
Figure showing the binding site residues. Ligands are displayed as
CPK. Figures were drawn with
Molscript (7) and rendered with
Raster3D (8). PISA coordinates
(3) are used when available
(all entries except NMR). Ligands do not appear on the picture when
PISA fails to apply symmetry operations to ligand coordinates. | | (click anywhere in this window to remove it) |
|
 |
|
List of binding site residues detected in this holo-structure. Column 1 gives the position, coloured on a yellow-to-red scale depending on the number of holo-structures where the residue is in contact with a ligand.
Column 2 gives the identifier of the chain to which the residue belongs.
Column 3 gives the 3-letter amino acid code, coloured according to physico-chemical type. The binding residues in this holo structure are listed for each ligand independently, following the ligand's unique ID. Note that since PISA files are used whenever available, chain identifiers may differ from those in original PDB files. | | (click anywhere in this window to remove it) |
|
| PO4 N2111 |
|---|
| 1026 | M |
GLY |
| 1030 | M |
ARG |
| 1091 | M |
ARG |
| 1093 | M |
GLY |
| 1094 | M |
THR |
| 1196 | M |
GLU |
| 3048 | R |
ARG |
| DUR G2102 |
|---|
| 1007 | H |
PHE |
| 1008 | H |
HIS |
| 1048 | H |
ARG |
| 1068 | E |
GLY |
| 1069 | E |
ILE |
| 1076 | H |
ILE |
| 1094 | E |
THR |
| 1095 | E |
THR |
| 1096 | E |
GLY |
| 1162 | E |
PHE |
| 1166 | E |
GLN |
| 1168 | E |
ARG |
| 1195 | E |
TYR |
| 1196 | E |
GLU |
| 1197 | E |
MET |
| 1198 | E |
GLU |
| | 1220 | E |
ILE |
| 1221 | E |
VAL |
| 1223 | E |
ARG |
| 1229 | E |
PRO |
| DUR L2122 |
|---|
| 1007 | E |
PHE |
| 1008 | E |
HIS |
| 1048 | E |
ARG |
| 1069 | H |
ILE |
| 1076 | E |
ILE |
| 1094 | H |
THR |
| 1095 | H |
THR |
| 1096 | H |
GLY |
| 1162 | H |
PHE |
| 1166 | H |
GLN |
| 1168 | H |
ARG |
| 1195 | H |
TYR |
| 1196 | H |
GLU |
| 1197 | H |
MET |
| 1198 | H |
GLU |
| 1220 | H |
ILE |
| 1221 | H |
VAL |
| 1223 | H |
ARG |
| 1229 | H |
PRO |
| | PO4 F2101 |
|---|
| 1026 | E |
GLY |
| 1030 | E |
ARG |
| 1048 | H |
ARG |
| 1091 | E |
ARG |
| 1092 | E |
ILE |
| 1093 | E |
GLY |
| 1094 | E |
THR |
| 1196 | E |
GLU |
| PO4 I2121 |
|---|
| 1026 | H |
GLY |
| 1030 | H |
ARG |
| 1048 | E |
ARG |
| 1091 | H |
ARG |
| 1093 | H |
GLY |
| 1094 | H |
THR |
| 1196 | H |
GLU |
| DUR Q2112 |
|---|
| 1069 | M |
ILE |
| 1094 | M |
THR |
| 1095 | M |
THR |
| 1096 | M |
GLY |
| 1162 | M |
PHE |
| 1166 | M |
GLN |
| 1168 | M |
ARG |
| | 1195 | M |
TYR |
| 1196 | M |
GLU |
| 1197 | M |
MET |
| 1198 | M |
GLU |
| 1220 | M |
ILE |
| 1221 | M |
VAL |
| 1223 | M |
ARG |
| 1229 | M |
PRO |
| 3007 | R |
PHE |
| 3008 | R |
HIS |
| 3048 | R |
ARG |
| 3076 | R |
ILE |
| PO4 B3111 |
|---|
| 2026 | A |
GLY |
| 2027 | A |
ASP |
| 2030 | A |
ARG |
| 2091 | A |
ARG |
| 2092 | A |
ILE |
| 2093 | A |
GLY |
| 2094 | A |
THR |
| 2198 | A |
GLU |
| 3048 | D |
ARG |
| DUR C3112 |
|---|
| 2069 | A |
ILE |
| 2094 | A |
THR |
| | 2095 | A |
THR |
| 2096 | A |
GLY |
| 2162 | A |
PHE |
| 2166 | A |
GLN |
| 2168 | A |
ARG |
| 2195 | A |
TYR |
| 2196 | A |
GLU |
| 2197 | A |
MET |
| 2198 | A |
GLU |
| 2220 | A |
ILE |
| 2221 | A |
VAL |
| 2223 | A |
ARG |
| 2229 | A |
PRO |
| 3007 | D |
PHE |
| 3008 | D |
HIS |
| 3048 | D |
ARG |
|
|
|
| PDB |
The Protein Data Bank |
| CSA |
Catalytic Site Atlas |
| PDBSum |
Overview of the macromolecular structure |
| CATH |
Protein Structure Classification |
| Scop |
Structural Classification of Proteins |
| Pfam |
Protein Families and Domains |
| UniProt |
Universal Protein Resource |
LIGPLOT (only on holo-pages) is hosted at the EBI. The LigPlot Jmol links point directly to the Jmol visualisation interface provided on the PDBSum page. Note that due to different software used, the atomic contacts of LigPlot and LigASite do not necessarily correspond. | | (click anywhere in this window to remove it) |
|
Links to external databases: LigPlot (hosted at the EBI):
|
|
Several files are provided for download: | • The XML file defining the residue-ligand contacts; this file contains data on the apo and all holo-structures. |
| • The XML Schema file defining the semantics of the XML file |
| • 3D coordinates of the structure used in constructing LigASite (PISA structure file whenever available, PDB file otherwise. |
| • 3D coordinates of the combined binding residues in the apo structure |
| • 3D coordinates of the binding residues of the holo structure (only on the holo page) |
Coordinate files are in PDB format. | | (click anywhere in this window to remove it) |
|
|
|
|
|
List of related structure, containing both the apo-structure
and other holo-structures.
Column 1 gives the PDB ID and column 2 the unique ID
of the ligands (holo-structures only).
Clicking the blue 'Hide table of related structures' button
removes the entire table. | | (click anywhere in this window to remove it) |
|
|
| pdb ID |
|---|
| 1t0u |
|
Details |
|
| pdb ID |
Ligand Unique ID |
|---|
| 1rxu |
PO4K2101 THML2102 |
Details |
|
PO4Q2121 THMR2122 |
|
PO4N2111 THMO2112 |
|
PO4H2091 THMI2092 |
|
PO4E2081 THMF2082 |
|
PO4B2071 THMC2072 |
| 1rxc |
R1PL2082 URFP2081 |
Details |
|
R1PF2032 URFJ2031 |
|
R1PD2012 URFB2011 |
|
R1PI2002 URFG2001 |
|
R1PO2022 URFM2021 |
| 3kvv |
R2BW_256 URFX_254 SO4V_255 |
Details |
|
R2BS_256 URFT_254 SO4R_255 |
|
R2BG_256 URFH_254 SO4F_255 |
|
R2BC_256 SO4B_255 URFD_254 |
|
R2BO_256 SO4N_255 URFP_254 |
|
R2BK_256 URFL_254 SO4J_255 |
|
|
|
Ligands present in this holo structure.
Column 1 shows the ligand HET code
Column 2 shows the name, chemical formula and (non-stereo) SMILES string.
Data in column 2 appears as 'not_found' when it is not present in the file
'pdb2smiles.xml' from www.rcsb.org. | | (click anywhere in this window to remove it) |
|
| DUR |
NAME: |
2'-DEOXYURIDINE |
|
FORMULA: |
C9 H12 N2 O5 |
|
SMILES: |
OCC1OC(CC1O)N2C=CC(=O)NC2=O |
| PO4 |
NAME: |
PHOSPHATE ION |
|
FORMULA: |
O4 P1 |
|
SMILES: |
[O-]P([O-])([O-])=O |
|
|
|
|
v9.7
April 2012 |
Interdisciplinary Research Institute, Computational Biology, Villeneuve d'Ascq, France
University College London, Biomolecular Structure and Modelling Unit, London, UK
Hospital for Sick Children and University of Toronto, Structural Biology and Biochemistry Program, Toronto, Canada |
| Script execution time: 0.6986 seconds |

